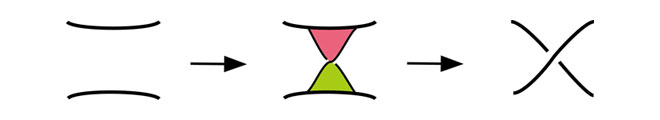
Knot theory is a branch of topology that deals with mathematical knots and links. A knot is an embedding of a circle into the 3-dimensional space. A union of knots is called a link or a catenane. As the chromosome of E. coli is closed circular, the core axis of its double stranded DNA can be considered as a knot. A site-specific recombination of DNA can be modeled mathematically by using a band surgery, a local move on a knot or a link as in the figure below. In biological experiments it is observed that some site-specific recombination reactions change the topology of DNA. Using this topological information, we can characterize the mechanisms of the site-specific recombination reactions as applications of results on band surgeries.
In our recent study, applying new mathematical results we characterized unlinking of replicated chromosome of E. coli by the FtsK-dependent XerC/XerD-dif recombination. There we determined the number of recombination steps needed to unlink replication catenanes. We showed that the stepwise unlinking as in the figure is the only possible pathway if we assume that the crossing number of DNA knot or link is reduced at each recombination. We also characterized the topological mechanism for these unlinking reactions.
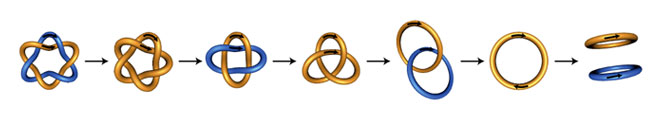
Reference: “FtsK-dependent XerCD-dif recombination unlinks replication catenanes in a stepwise manner”, Koya Shimokawa, Kai Ishihara, Ian Grainge, David J. Sherratt, and Mariel Vazquez, Proc. Natl. Acad. Sci. U S A (in press).


© Copyright Saitama University, All Rights Reserved.